Pathways /
RNA splicing
Overview
The process of eukaryotic RNA splicing involves the excision of non-coding introns and the preservation and joining of coding exons in an RNA molecule. This process is achieved via the use of a spliceosome. RNA splicing is activated by the binding of the spliceosome to the RNA molecule. [1]
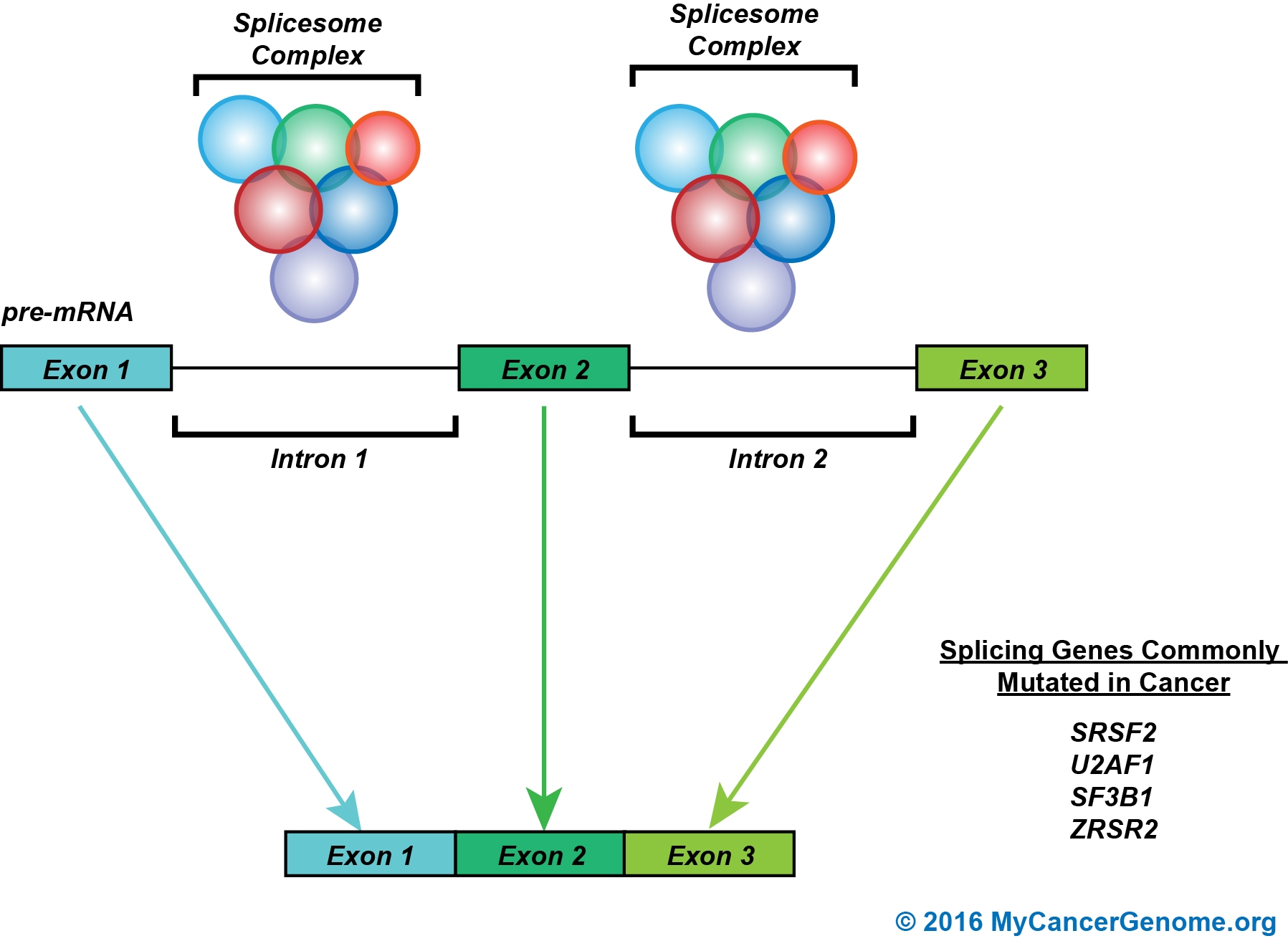
Figure 1. RNA splicing is the process whereby non-coding introns are removed to create a sequence of coding exons. The splicing process is mediated by the spliceosome. The spliceosome is a complex of several RNA protein molecules including but not limited to ZRSR2, SRSF2, SF3B1, and U2AF1. Broadly, the spliceosome removes intronic sequences to generate the mRNA which will be used to translate the resultant protein. Specific nodes in the pathway that are therapeutically actionable are noted.

 Back to Pathways List
Back to Pathways List