About
-
About
-
Molecular Medicine
My Cancer Genome Data Sources
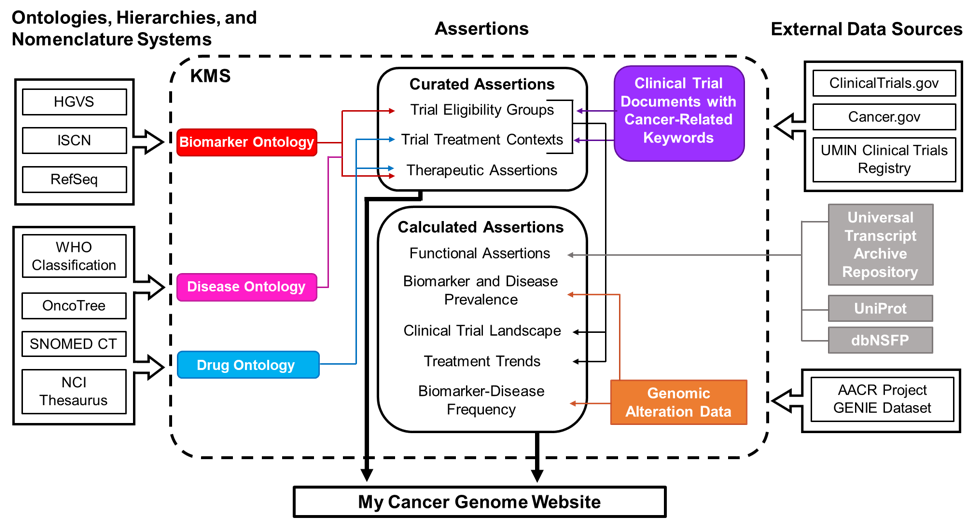
Figure 1. Schematic of data sources for My Cancer Genome. Curated assertions on the My Cancer Genome website are housed within a Knowledge Management System (KMS) created through a partnership with GenomOncology that serves as a warehouse of biomarker, disease, and drug ontologies; biomarker-driven cancer clinical trials; therapeutic, prognostic, and diagnostic assertions; and unstructured content describing pathways, molecular testing techniques, and terms related to precision oncology. Ontologies are developed using publicly available nomenclatures, hierarchies, and ontologies, such as the NCI Thesaurus, and manually supplemented as necessary. Cancer clinical trial documents are imported from external document sources into the KMS, where biomarker-related eligibility criteria and therapeutic interventions are annotated to generate a database of curated clinical trial eligibility groups and treatment contexts. Therapeutic assertions describe the predicted response to a drug in the context of a specific disease setting and biomarker status. These assertions are manually curated from drug approvals, treatment guidelines, and literature review prior to release to the My Cancer Genome website. Biomarker and disease frequency data displayed in the Overview and Significance sections of biomarker and disease pages are derived from the publicly available AACR Project GENIE genomic alteration dataset.
Biomarkers, diseases, and drugs highlighted on the My Cancer Genome website are associated with therapeutic assertions and curated clinical trials in the KMS. The website also displays detail pages for biomarkers associated with five or more GENIE cases.
Definitions and Related Publications
The KMS and our clinical trial curation process is described in more detail in the following manuscript: Jain et al. Detailed data model for accurate curation of clinical trial eligibility criteria and an analysis of the current clinical trial landscape. In preparation.
AACR Project GENIE is described in more detail in the following manuscripts:
- The AACR Project GENIE Consortium. AACR Project GENIE: powering precision medicine through an international consortium. Cancer Discovery. 2017;7(8):818-831. PMID: 28572459
- Micheel CM, Sweeney SM, LeNoue-Newton ML, et al. American Association for Cancer Research Project Genomics Evidence Neoplasia Information Exchange: from inception to first data release and beyond–lessons learned and member institutions’ perspectives. JCO Clinical Cancer Informatics. 2018;2:1-14. PMID: 30652542
The National Comprehensive Cancer Network (NCCN) guidelines are referenced with permission from the NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®). © National Comprehensive Cancer Network, Inc. 2019. All rights reserved. To view the most recent and complete version of the guideline, go online to NCCN.org. NCCN makes no warranties of any kind whatsoever regarding their content, use or application and disclaims any responsibility for their application or use in any way.
For more information about data sources for the KMS, please visit the Glossary:
Cancer.gov
ClinicalTrials.gov
dbNSFP
HGVS
ISCN
NCI Thesaurus
OncoTree
RefSeq
SNOWMED CT
WHO Classification
UniProt
Universal Transcript Archive
UMIN Clinical Trials Registry
For more information about the My Cancer Genome curation process or the GenomOncology software tools, please see the APIs and Licensing page.
Last updated: Nov 12, 2019
Disclaimer: The information presented at MyCancerGenome.org is compiled from sources believed to be reliable. Extensive efforts have been made to make this information as accurate and as up-to-date as possible. However, the accuracy and completeness of this information cannot be guaranteed. Despite our best efforts, this information may contain typographical errors and omissions. The contents are to be used only as a guide, and health care providers should employ sound clinical judgment in interpreting this information for individual patient care.